Tune in to the latest #eLifePodcast to hear authors discuss the stories behind their recent discoveries https://t.co/awwqu5L3lE
eLife - the journal (@eLife)
Altmetric Small
Altmetric provides a collated score for online attention across various platforms and media.
See more details
Caption Text
[Do not use this base pattern directly: see the following variants of this pattern for usage]
Caption Text Additional Asset
Table 1 - source data 1.
Is the species name mentioned in the title, impact statement, abstract and digest of eLife papers (July–Sept 2014)?
Caption Text Captioned Asset
Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
Listed are, for each triplet of cell types, the probabilities of the four topologies for prior odds ; the number of topologies that reach probability for some value of between 10−6 and 102; the non-null topology that has the highest probability over the range of prior odds (if the null topology is the most likely topology for the entire range of prior odds, the topology is marked ‘null’); and the value of highest probability over the range of prior odds; the correct topology and triplet length in the traditional model; and the correct topology and triplet length in the Adolfsson model.
(A) Doxycycline-inducible gam-gfp fusion construct in the E. coli chromosome. Constitutively produced TetR protein represses the PN25tetO promoter, which produces GamGFP upon doxycycline induction. oriC, origin of replication; ter, replication terminus; arrows, directions of transcription. (B) Phage λ assay for end-blocking activity by Mu Gam and GamGFP. Rolling-circle replication of phage λred gam is inhibited by E. coli RecBCD, which causes small plaques of λred gam on wild-type E. coli (Smith, 1983). Mu Gam protein binds and protects DNA ends from RecBCD exonuclease activity (Akroyd and Symonds, 1986) and so is expected to allow rolling-circle replication of λred gam and therefore allow formation of large plaques. (C) λred gam plaques are small on recB+ (WT) and large on recB-deficient cells (recB-). Plaques produced on WT cells carrying gam and gam-gfp are small when Gam and GamGFP proteins are not produced (Uninduced). (D) λred gam produce large plaques on WT cells if Gam or GamGFP are produced (Induced). (E) UV sensitivity of E. coli recB-null mutant compared with recB+(WT), and uninduced gam and gam-gfp carrying cells. WT ( ), recB− (
), recB− ( ), WT GamGFP, (
), WT GamGFP, ( ); WT Gam, (
); WT Gam, ( ). (F) Induction of Gam or GamGFP with 200 ng/ml doxycycline causes UV sensitivity similar to that of recB-null mutant cells, indicating that Gam or GamGFP block RecBCD action on double-stranded DNA ends. WT, SMR14327; recB, SMR8350; WT GamGFP, SMR14334; WT Gam, SMR14333. Representative experiment performed three times with comparable results.
). (F) Induction of Gam or GamGFP with 200 ng/ml doxycycline causes UV sensitivity similar to that of recB-null mutant cells, indicating that Gam or GamGFP block RecBCD action on double-stranded DNA ends. WT, SMR14327; recB, SMR8350; WT GamGFP, SMR14334; WT Gam, SMR14333. Representative experiment performed three times with comparable results.
Code
<dock_design>
<SCOREFXNS>
<fullatom weights=beta symmetric = 0> </fullatom>
</SCOREFXNS>
<!-- Longabadaliasametatquedoloreaqueesthicillabadaliasametatquedoloreaqueesthicillabadaliasametatquedoloreaqueesthicillolong -->
<FILTERS>
<Ddg name=ddg scorefxn=fullatom threshold = 0 jump = 1 repeats = 1 repack = 1 confidence = 1/>
<Sasa name=sasa confidence = 0/>
<ShapeComplementarity name=shape verbose = 1 confidence = 0 jump = 1/>
</FILTERS>
<MOVERS>
<AtomTree name=docking_tree docking_ft = 1/>
<DockSetupMover name=setup_dock/>
<DockingProtocol name=dock docking_score_high=fullatom low_res_protocol_only = 0 docking_local_refine = 0 dock_min = 1 />
</MOVERS>
<APPLY_TO_POSE>
</APPLY_TO_POSE>
<PROTOCOLS>
<Add mover_name=docking_tree/>
<Add mover_name=setup_dock/>
<Add mover_name=dock/>
<Add filter_name=ddg/>
<Add filter_name=sasa/>
<Add filter_name=shape/>
</PROTOCOLS>
</dock_design>Date Updated
Definition List
- Research focus
- Pre-mRNA splicing
- post-transcriptional gene regulation
- Experimental organism
- Human
- Mouse
Definition List Inline
- Research focus
- Pre-mRNA splicing
- post-transcriptional gene regulation
- Experimental organism
- Human
- Mouse
Definition List Timeline With Version
- Version of Record
- Read the peer reviews
- Reviewed Preprint
- v3
- Reviewed Preprint
- v2
- Reviewed Preprint
- v1
Definition List Timeline Without Version
- Version of Record
- Accepted Manuscript
Doi Without Link
List Heading
Podcasts
List
- item 1
- item 2
List Bullet
- item 1
- item 2
List Line
- Version of Record published
- Accepted Manuscript published
- Accepted
- Received
- Preprint posted
List Numbers
- item 1
- item 2
List Roman
- item 1
- item 2
Math Long
Math With Label
Paragraph
Some text in a paragraph element
Previous Version Warning
Process Block New Process
Version of Record: This is the final version of the article.
Read more about eLife's peer review process.Table
| F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | Partial η2 | Original effect size f | Replication total sample size | Detectable effect size f |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F(24,39) = 0.8678 (interaction) | 0.348120 | 0.7307699 | 169* | 0.3895070 | ||||||||
| F(2,39) = 0.8075 (treatments) | 0.039766 | 0.2035014 | 169 | 0.2415459 | ||||||||
| F(12,39) = 187.6811 (hematology parameters) | 0.982978 | 7.599178 | 169* | 0.3331365 |
| F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | Partial η2 | Original effect size f | Replication total sample size | Detectable effect size f |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F(24,39) = 0.8678 (interaction) | 0.348120 | 0.7307699 | 169* | 0.3895070 | ||||||||
| F(2,39) = 0.8075 (treatments) | 0.039766 | 0.2035014 | 169* | 0.2415459 | ||||||||
| F(12,39) = 187.6811 (hematology parameters) | 0.982978 | 7.599178 | 169* | 0.3331365 |
-
*
Footnote 1.
-
The values in parenthesis refer to the highest resolution shell.
* where is the intensity of an individual measurement of a reflection and is the average intensity of that reflection.
Table Code Blocks
| data = {{},{},{}...,{}}; loglog=Function[{x,y},{Log[x/],Log[y]}]; datalog=Apply[loglog,data,{1}]; peakr = Function[{},Piecewise[{{+(1/2)*Log[1-(2/3)*] +*(1-(1-(2/3)*)^(3/2),<3/2/,Infinity}}]]; fit = NonlinearModelFit[datalog,{peakr[],{}}, {{},{},{}},x,Method->NMinimize]; fit['ParameterTable'] |
-
Table with Code Blocks
Table Equations
| Definitions of the simulated system | |
|---|---|
| Type | Status |
| Protein assembly occupancy | |
| Histones | |
| Regions of histones | |
-
Table with Equations
Table Gene Sequences
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Recombinant DNA reagent | synthesized gene - mScarletI | Genscript | actagtggtggttcaggaGTTTCAAAAGGTGAAGCCGTTATTAAAGAATTTATGAGATTCAAGGTTCACATGGAAGGAAGTATGAACGGTCATGAATTTGAGATTGAAGGAGAAGGTGAAGGTAGACCATATGAAGGCACCCAAACAGCTAAATTAAAAGTAACTAAAGGTGGTCCATTACCATTTAGTTGGGATATTTTATCTCCACAATTTATGTATGGTTCACGTGCTTTCAttAAACATCCAGCAGATATTCCAGATTATTATAAACAATCATTTCCAGAAGGTTTTAAATGGGAACGTGTCATGAACTTTGAAGATGGTGGAGCAGTTACAGTCACACAAGATACCTCATTAGAAGATGGTACATTAATATATAAAGTTAAATTACGTGGTACTAATTTTCCACCAGACGGTCCAGTAATGCAAAAAAAAACAATGGGCTGGGAAGCTAGTACAGAACGTTTATATCCTGAAGATGGTGTCCTTAAAGGCGATATAAAAATGGCCTTGAGATTAAAGGATGGTGGTAGGTATTTAGCAGATTTCAAAACCACTTATAAAGCAAAAAAACCAGTTCAAATGCCAGGTGCATATAATGTTGATAGAAAACTTGATATTACCAGTCATAATGAAGATTACACAGTTGTCGAACAATACGAACGTTCTGAAGGTCGTCATAGCACTGGTGGTATGGATGAATTATACAAATAAgctagc | d |
| Recombinant DNA reagent | synthesized gene - mNeon | Genscript | actagtggtggttcaggaGTTTCAAAAGGTGAAGCCGTTATTAAAGAATTTATGAGATTCAAGGTTCACATGGAAGGAAGTATGAACGGTCATGAATTTGAGATTGAAGGAGAAGGTGAAGGTAGACCATATGAAGGCACCCAAACAGCTAAATTAAAAGTAACTAAAGGTGGTCCATTACCATTTAGTTGGGATATTTTATCTCCACAATTTATGTATGGTTCACGTGCTTTCAttAAACATCCAGCAGATATTCCAGATTATTATAAACAATCATTTCCAGAAGGTTTTAAATGGGAACGTGTCATGAACTTTGAAGATGGTGGAGCAGTTACAGTCACACAAGATACCTCATTAGAAGATGGTACATTAATATATAAAGTTAAATTACGTGGTACTAATTTTCCACCAGACGGTCCAGTAATGCAAAAAAAAACAATGGGCTGGGAAGCTAGTACAGAACGTTTATATCCTGAAGATGGTGTCCTTAAAGGCGATATAAAAATGGCCTTGAGATTAAAGGATGGTGGTAGGTATTTAGCAGATTTCAAAACCACTTATAAAGCAAAAAAACCAGTTCAAATGCCAGGTGCATATAATGTTGATAGAAAACTTGATATTACCAGTCATAATGAAGATTACACAGTTGTCGAACAATACGAACGTTCTGAAGGTCGTCATAGCACTGGTGGTATGGATGAATTATACAAATAAgctagc |
-
Table with Gene Sequences
Table Inline Images
| unwinding | turnover | ||
|---|---|---|---|
| # | Chemical drawing | IC50[95%CI];µM | IC50[95%CI];µM |
| 2 | 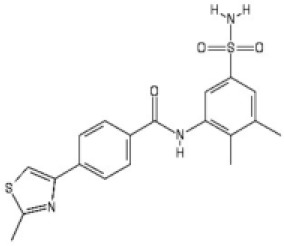 | 2.2 [1.7–2.7] | 3.2 [2.3–4.0] |
-
Table with inline images
Table Long Url
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Software, algorithm | Metamorph | Metamorph v | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy#gref |
-
Table with long url
Table Ordered List
| Factors classically associated with immune functions | ||||
|---|---|---|---|---|
| Protein | Immune system properties | Nervous system properties | References | |
| Antimicrobial peptides(AMPs) |
|
| Hanson et al., 2019;Lezi et al., 2018; | |
-
Table with ordered list
Table Unordered List
| Factors classically associated with immune functions | ||||
|---|---|---|---|---|
| Protein | Immune system properties | Nervous system properties | References | |
| Antimicrobial peptides(AMPs) |
|
| Hanson et al., 2019;Lezi et al., 2018;Su et al., 2010;Zasloff, 2002 | |
-
Table with unordered list
Term
Term Highlighted
Tweet
Tweet With Cards
Tune in to the latest #eLifePodcast to hear authors discuss the stories behind their recent discoveries https://t.co/awwqu5L3lE
eLife - the journal (@eLife)
Client Error
Say what?
Your browser sent us something unexpected.
We’ve logged the problem and will look into it.
Not Found

Oops!
The page you were looking for is not found.
Orcid Error Visibility Setting

Your ORCID iD settings prevented log in
Please update the visibility settings of the name associated with your ORCID iD before trying again.
Step 1: Open your name's visibility settings on your ORCID iD page
Go to orcid.org/my-orcid to change the visibility settings of your name by using the pencil icon.
It should be in the top left corner of your ORCID iD page, next to your name.

Step 2: Change the visibility setting
Change the visibility setting to "everyone" or "trusted parties" and save your changes. Locking your name will not allow you to log in to eLife.
You may be asked to authorise eLife as a trusted party when you try to log in again. Learn more about ORCID visibility settings.
Step 3: Return to eLife and log in again
Return to elifesciences.org and attempt to log in again. If you are still unable to log in please contact us.

Server Error
Kabooooooom!
Something has gone seriously wrong.
We’ve logged the problem and will look into it.
Checkbox
Checkboxes
-
Children detected!
Checkboxes Disabled
-
Children detected!
Checkboxes Error
-
Children detected!
Checkboxes Info Error
-
Children detected!
Checkboxes Info
-
Children detected!
Reference
Reference Book Chapter
Reference Book
Reference Clinical Trial
Reference Conference With Doi
Reference Conference With Link
Reference Conference
Reference Data
Reference Journal Article With Doi Without Link
Reference Patent
Reference Software
Reference Thesis
Reference Web
Article Assessment Terms
- Landmark
- Fundamental
- Important
- Valuable
- Useful
About Profile
[Do not use this base pattern directly: see the following variants of this pattern for usage]
About Profile Image

Randy Scheckman
Editor-in-Chief
Lorem ipsum dolor sit amet, consectetur adipiscing elit. Mauris tempor euismod mi. Morbi facilisis eros in mi consequat cursus. Nulla malesuada pretium mauris, vitae gravida diam laoreet ac. Curabitur ornare libero sed porta molestie. Ut ut sem in libero tincidunt lobortis. Nullam eros nisi, malesuada vitae leo sit amet, bibendum congue justo. Morbi iaculis lectus erat, vel dapibus metus posuere sit amet. Phasellus eget scelerisque massa. Mauris hendrerit ex in nisl ultricies elementum. Donec tellus risus, rhoncus molestie rutrum et, cursus eget mi. Praesent et elit tellus. Curabitur id sodales turpis. Fusce pulvinar aliquam commodo. Cras vitae dolor vitae metus dignissim auctor.Cras nulla purus, rhoncus et mollis in, imperdiet sed metus. In non porttitor nibh. In sit amet ipsum eget nisl laoreet tincidunt. Donec iaculis nibh quis lacus viverra sagittis. Phasellus imperdiet congue imperdiet.
Interdum et malesuada fames ac ante ipsum primis in faucibus. Nunc nisi tortor, rhoncus.
- Expertise
- Cell Biology
- Competing interests statement
- Donec tellus
About Profile No Image
Randy Scheckman
Editor-in-Chief
Lorem ipsum dolor sit amet, consectetur adipiscing elit. Mauris tempor euismod mi. Morbi facilisis eros in mi consequat cursus. Nulla malesuada pretium mauris, vitae gravida diam laoreet ac. Curabitur ornare libero sed porta molestie. Ut ut sem in libero tincidunt lobortis. Nullam eros nisi, malesuada vitae leo sit amet, bibendum congue justo. Morbi iaculis lectus erat, vel dapibus metus posuere sit amet. Phasellus eget scelerisque massa. Mauris hendrerit ex in nisl ultricies elementum. Donec tellus risus, rhoncus molestie rutrum et, cursus eget mi. Praesent et elit tellus. Curabitur id sodales turpis. Fusce pulvinar aliquam commodo. Cras vitae dolor vitae metus dignissim auctor.Cras nulla purus, rhoncus et mollis in, imperdiet sed metus. In non porttitor nibh. In sit amet ipsum eget nisl laoreet tincidunt. Donec iaculis nibh quis lacus viverra sagittis. Phasellus imperdiet congue imperdiet.
Interdum et malesuada fames ac ante ipsum primis in faucibus. Nunc nisi tortor, rhoncus.
- Expertise
- Cell Biology
- Competing interests statement
- Donec tellus
About Profile Placeholder Image

Randy Scheckman
Editor-in-Chief
Lorem ipsum dolor sit amet, consectetur adipiscing elit. Mauris tempor euismod mi. Morbi facilisis eros in mi consequat cursus. Nulla malesuada pretium mauris, vitae gravida diam laoreet ac. Curabitur ornare libero sed porta molestie. Ut ut sem in libero tincidunt lobortis. Nullam eros nisi, malesuada vitae leo sit amet, bibendum congue justo. Morbi iaculis lectus erat, vel dapibus metus posuere sit amet. Phasellus eget scelerisque massa. Mauris hendrerit ex in nisl ultricies elementum. Donec tellus risus, rhoncus molestie rutrum et, cursus eget mi. Praesent et elit tellus. Curabitur id sodales turpis. Fusce pulvinar aliquam commodo. Cras vitae dolor vitae metus dignissim auctor.Cras nulla purus, rhoncus et mollis in, imperdiet sed metus. In non porttitor nibh. In sit amet ipsum eget nisl laoreet tincidunt. Donec iaculis nibh quis lacus viverra sagittis. Phasellus imperdiet congue imperdiet.
Interdum et malesuada fames ac ante ipsum primis in faucibus. Nunc nisi tortor, rhoncus.
- Expertise
- Cell Biology
- Competing interests statement
- Donec tellus
Additional Asset
[Do not use this base pattern directly: see the following variants of this pattern for usage]
Additional Asset Doi
Table 1 - source data 1.
Is the species name mentioned in the title, impact statement, abstract and digest of eLife papers (July–Sept 2014)?
Additional Asset Non Doi
Table 1 - source data 1.
Is the species name mentioned in the title, impact statement, abstract and digest of eLife papers (July–Sept 2014)?
Article Download Links List
Downloads (links to download the article or parts of the article as PDF, and in DAR format)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Article Section Kitchen Sink
The Kitchen Sink
Some linkHeading 1
Heading 2
Heading 3
Heading 4
Heading 5
Heading 6
This is not wrapped in a paragraph.
This is in a paragraph. Lorem ipsum dolor sit amet, test link adipiscing elit. This is strong. Nullam dignissim convallis est. This is underlined. Quisque aliquam. This is emphasized. Donec faucibus. This is monospace. Nunc iaculis suscipit dui. 53 = 125. Water is H2O. Nam sit amet sem. This is in small caps. Aliquam libero nisi, imperdiet at, tincidunt nec, gravida vehicula, nisl. The New York Times (That’s a citation). Maecenas ornare tortor. Donec sed tellus eget sapien fringilla nonummy. Mauris a ante. Suspendisse quam sem, consequat at, commodo vitae, feugiat in, nunc. Morbi imperdiet augue quis tellus.
HTML and CSS are our tools. Mauris a ante. Suspendisse quam sem, consequat at, commodo vitae, TAATAAGGAAGAACTGCTTATTCTTAATTATTTCTACCTACTAAACTAACTAATTATCAACAAATATCATCTATTTAATAGTATATCATCACATGCGGTGTAAGAGGATGACATAAAGATTGAGAAACAGTCATCCAGTCTAATGGAAGCTCAAATGCAAGGGCTGATAATGTAATAGGATAATGAATGACAACGTATAAAAGGAAAGAAGATAAAGCAATATTATTTTGTAGAATTATCGATTCCCTTTTGTGGATCCCTATATCCTCGAGGAGAA feugiat in, nunc. Morbi imperdiet augue quis tellus. Praesent mattis, massa quis luctus fermentum, turpis mi volutpat justo, eu volutpat enim diam eget metus. To copy a file type COPY filename. Dinner’s at 5:00. Let’s make that 7. This text has been struck.
List Types
Definition List
- Definition List Title
- This is a definition list division.
- Definition
- An exact statement or description of the nature, scope, or meaning of something: our definition of what constitutes poetry.
Ordered List
- List Item 1
- List Item 2
- Nested list item A
- Nested list item B
- List Item 3
Unordered List
- List Item 1
- List Item 2
- Nested list item A
- Nested list item B
- List Item 3
Table
| Table Header 1 | Table Header 2 | Table Header 3 | Table Header 4 |
|---|---|---|---|
| Division 1 | Division 2 | Division 3 | Division 4 |
| Division 1 | Division 2 | Division 3 | Division 4 |
| Division 1 | Division 2 | Division 3 with link | Division 4 with link |
| This is blue. | This is green. | This is orange. | This is yellow. |
| This is purple. | This is red. | This is pink. | This is grey. |
Preformatted Text
Typographically, preformatted text is not the same thing as code. Sometimes, a faithful execution of the text requires preformatted text that may not have anything to do with code. Most browsers use Courier and that’s a good default — with one slight adjustment, Courier 10 Pitch over regular Courier for Linux users. For example:
“Beware the Jabberwock, my son! The jaws that bite, the claws that catch! Beware the Jubjub bird, and shun The frumious Bandersnatch!”
Code
Code can be presented inline, like <?php bloginfo('stylesheet_url'); ?>, or within a <pre> block. Because we have more specific typographic needs for code, we’ll specify Consolas and Monaco ahead of the browser-defined monospace font.
#container {
float: left;
margin: 0 -240px 0 0;
width: 100%;
}
Blockquotes
Let’s keep it simple. Italics are good to help set it off from the body text. Be sure to style the citation.
Good afternoon, gentlemen. I am a HAL 9000 computer. I became operational at the H.A.L. plant in Urbana, Illinois on the 12th of January 1992. My instructor was Mr. Langley, and he taught me to sing a song. If you’d like to hear it I can sing it for you.
— Citing HAL 9000
And here’s a bit of trailing text.
Article Section
Article Section Assessment
Assessment
This valuable paper informs on the role of type I PRMTs in programming muscle stem cell identification. The evidence presented is mostly solid, with some weaknesses in the evidence regarding the proposed mechanism. The paper will be of particular interest to those who study skeletal muscle satellite cell biology.
https://doi.org/10.7554/eLife.62927.sa1- Landmark
- Fundamental
- Important
- Valuable
- Useful
- Exceptional
- Compelling
- Convincing
- Solid
- Incomplete
- Inadequate
During the peer-review process the editor and reviewers write an eLife Assessment that summarises the significance of the findings reported in the article (on a scale ranging from landmark to useful) and the strength of the evidence (on a scale ranging from exceptional to inadequate). Learn more about eLife Assessments
Article Section Default
Introduction
TNF Receptor Associated Factor 2 (TRAF2) is an adaptor protein that transduces signals following ligation of certain cytokine receptors including those binding TNF. It was first identified together with TRAF1 as a component of TNF receptor-2 and then TNF receptor-1 (TNFR1) signalling complexes (Rothe et al., 1994; Shu et al., 1996). TRAF2, like most other TRAFs, contains a RING domain, several zinc fingers, a TRAF-N, and a conserved TRAF-C domain which is responsible for oligomerisation and receptor binding through its MATH region (Takeuchi et al., 1996; Uren and Vaux, 1996).
RING domains are nearly always associated with ubiquitin E3 ligase activity (Shi and Kehrl, 2003) and TRAF2 can promote ubiquitylation of RIPK1 in TNFR1 signalling complexes (TNFR1-SC) (Wertz et al., 2004). However TRAF2 recruits E3 ligases such as cIAPs to TNFR1-SC and these have also been shown to be able to ubiquitylate RIPK1 and regulate TNF signalling (Dynek et al., 2010; Mahoney et al., 2008; Varfolomeev et al., 2008; Vince et al., 2009). This makes it difficult to unambiguously determine the role of the E3 ligase activity of TRAF2.
Activation of JNK and NF-κB by TNF is reduced in cells from Traf2-/- mice while only JNK signalling was affected in lymphocytes from transgenic mice that express a dominant negative (DN) form of TRAF2 that lacks the RING domain (Lee et al., 1997; Yeh et al., 1997). Traf2-/-Traf5-/- mouse embryonic fibroblasts (MEFs) have a pronounced defect in activation of NF-κB by TNF, suggesting that absence of TRAF2 can be compensated by TRAF5 (Tada et al., 2001). Although activation of NF-κB was restored in Traf2-/-Traf5-/- cells by re-expression of wild type TRAF2, it was not restored when the cells were reconstituted with TRAF2 point mutants that could not bind cIAPs (Vince et al., 2009; Zhang et al., 2010). These data, together with a wealth of different lines of evidence showing that cIAPs are critical E3 ligases required for TNF-induced canonical NF-κB (Blackwell et al., 2013; Haas et al., 2009; Silke, 2011), support the idea that the main function of TRAF2 in TNF-induced NF-κB is to recruit cIAPs to the TNFR1-SC. However, it remains possible that the RING of TRAF2 plays another function, such as in activating JNK and protecting cells from TNF-induced cell death (Vince et al., 2009; Zhang et al., 2010). Furthermore it has been shown that TRAF2 can K48-ubiquitylate caspase-8 to set the threshold for TRAIL or Fas induced cell death (Gonzalvez et al., 2012). Moreover, TRAF2 inhibits non-canonical NF-κB signalling (Grech et al., 2004; Zarnegar et al., 2008) and this function requires the RING domain of TRAF2 to induce proteosomal degradation of NIK (Vince et al., 2009). However, structural and in vitro analyses indicate that, unlike TRAF6, the RING domain of TRAF2 is unable to bind E2 conjugating enzymes (Yin et al., 2009), and is therefore unlikely to have intrinsic E3 ligase activity.
Sphingosine-1-phosphate (S1P) is a pleiotropic sphingolipid mediator that regulates proliferation, differentiation, cell trafficking and vascular development (Pitson, 2011). S1P is generated by sphingosine kinase 1 and 2 (SPHK1 and SPHK2) (Kohama et al., 1998; Liu et al., 2000). Extracellular S1P mainly acts by binding to its five G protein-coupled receptors S1P1-5 (Hla and Dannenberg, 2012). However, some intracellular roles have been suggested for S1P, including the blocking of the histone deacetylases, HDAC1/2 (Hait et al., 2009) and the induction of apoptosis through interaction with BAK and BAX (Chipuk et al., 2012).
Recently, it was suggested that the RING domain of TRAF2 requires S1P as a co-factor for its E3 ligase activity (Alvarez et al., 2010). Alvarez and colleagues proposed that SPHK1 but not SPHK2 is activated by TNF and phosphorylates sphingosine to S1P which in turn binds to the RING domain of TRAF2 and serves as an essential co-factor that was missing in the experiments of Yin et al. Alvarez and colleagues, observed that in the absence of SPHK1, TNF-induced NF-κB activation was completely abolished.
Although we know a lot about TRAF2, there are still important gaps particularly with regard to cell type specificity and in vivo function of TRAF2. Moreover, despite the claims that SPHK1 and its product, S1P, are required for TRAF2 to function as a ubiquitin ligase, the responses of Traf2-/- and Sphk1-/- cells to TNF were not compared. Therefore, we undertook an analysis of TRAF2 and SPHK1 function in TNF signalling in a number of different tissues.
Surprisingly, we found that neither TRAF2 nor SPHK1 are required for TNF mediated canonical NF-κB and MAPK signalling in macrophages. However, MEFs, murine dermal fibroblasts (MDFs) and keratinocytes required TRAF2 but not SPHK1 for full strength TNF signalling. In these cell types, absence of TRAF2 caused a delay in TNF-induced activation of NF-κB and MAPK, and sensitivity to killing by TNF was increased. Absence of TRAF2 in keratinocytes in vivo resulted in psoriasis-like epidermal hyperplasia and skin inflammation. Unlike TNF-dependent genetic inflammatory skin conditions, such as IKK2 epidermal knock-out (Pasparakis et al., 2002) and the cpdm mutant (Gerlach et al., 2011), the onset of inflammation was only delayed, and not prevented by deletion of TNF. This early TNF-dependent inflammation is caused by excessive apoptotic but not necroptotic cell death and could be prevented by deletion of Casp8. We observed constitutive activation of NIK and non-canonical NF-κB in Traf2-/- keratinocytes which caused production of inflammatory cytokines and chemokines. We were able to reverse this inflammatory phenotype by simultaneously deleting both Tnf and Nfkb2 genes. Our results highlight the important role TRAF2 plays to protect keratinocytes from cell death and to down-regulate inflammatory responses and support the idea that intrinsic defects in keratinocytes can initiate psoriasis-like skin inflammation.
https://doi.org/10.7554/eLife.16370Article Section Editor Evaluation
Editor's evaluation
The manuscript by Rosello et al. represents a major advance in implementation of cutting-edge genome editing methodologies in the zebrafish. The study seeks to describe optimized tools for precise base editing in zebrafish and to demonstrate their effective application. Overall, this study demonstrates that cytosine base editing is an efficient and powerful method for introducing precise in vivo edits into the zebrafish genome, and will be of interest to scientists who use zebrafish and other genetic systems to model human development and disease.
https://doi.org/10.7554/eLife.62927.sa1Article Section Editor Title Text
- Dominique Soldati-Favre
- University of Geneva, Switzerland
Article Section Header Link
TNF Receptor Associated Factor 2 (TRAF2) is an adaptor protein that transduces signals following ligation of certain cytokine receptors
some text to carry a linkTNF Receptor Associated Factor 2 (TRAF2) is an adaptor protein that transduces signals following ligation of certain cytokine receptors including those binding TNF. It was first identified together with TRAF1 as a component of TNF receptor-2 and then TNF receptor-1 (TNFR1) signalling complexes (Rothe et al., 1994; Shu et al., 1996). TRAF2, like most other TRAFs, contains a RING domain, several zinc fingers, a TRAF-N, and a conserved TRAF-C domain which is responsible for oligomerisation and receptor binding through its MATH region (Takeuchi et al., 1996; Uren and Vaux, 1996).
RING domains are nearly always associated with ubiquitin E3 ligase activity (Shi and Kehrl, 2003) and TRAF2 can promote ubiquitylation of RIPK1 in TNFR1 signalling complexes (TNFR1-SC) (Wertz et al., 2004). However TRAF2 recruits E3 ligases such as cIAPs to TNFR1-SC and these have also been shown to be able to ubiquitylate RIPK1 and regulate TNF signalling (Dynek et al., 2010; Mahoney et al., 2008; Varfolomeev et al., 2008; Vince et al., 2009). This makes it difficult to unambiguously determine the role of the E3 ligase activity of TRAF2.
Activation of JNK and NF-κB by TNF is reduced in cells from Traf2-/- mice while only JNK signalling was affected in lymphocytes from transgenic mice that express a dominant negative (DN) form of TRAF2 that lacks the RING domain (Lee et al., 1997; Yeh et al., 1997). Traf2-/-Traf5-/- mouse embryonic fibroblasts (MEFs) have a pronounced defect in activation of NF-κB by TNF, suggesting that absence of TRAF2 can be compensated by TRAF5 (Tada et al., 2001). Although activation of NF-κB was restored in Traf2-/-Traf5-/- cells by re-expression of wild type TRAF2, it was not restored when the cells were reconstituted with TRAF2 point mutants that could not bind cIAPs (Vince et al., 2009; Zhang et al., 2010). These data, together with a wealth of different lines of evidence showing that cIAPs are critical E3 ligases required for TNF-induced canonical NF-κB (Blackwell et al., 2013; Haas et al., 2009; Silke, 2011), support the idea that the main function of TRAF2 in TNF-induced NF-κB is to recruit cIAPs to the TNFR1-SC. However, it remains possible that the RING of TRAF2 plays another function, such as in activating JNK and protecting cells from TNF-induced cell death (Vince et al., 2009; Zhang et al., 2010). Furthermore it has been shown that TRAF2 can K48-ubiquitylate caspase-8 to set the threshold for TRAIL or Fas induced cell death (Gonzalvez et al., 2012). Moreover, TRAF2 inhibits non-canonical NF-κB signalling (Grech et al., 2004; Zarnegar et al., 2008) and this function requires the RING domain of TRAF2 to induce proteosomal degradation of NIK (Vince et al., 2009). However, structural and in vitro analyses indicate that, unlike TRAF6, the RING domain of TRAF2 is unable to bind E2 conjugating enzymes (Yin et al., 2009), and is therefore unlikely to have intrinsic E3 ligase activity.
Sphingosine-1-phosphate (S1P) is a pleiotropic sphingolipid mediator that regulates proliferation, differentiation, cell trafficking and vascular development (Pitson, 2011). S1P is generated by sphingosine kinase 1 and 2 (SPHK1 and SPHK2) (Kohama et al., 1998; Liu et al., 2000). Extracellular S1P mainly acts by binding to its five G protein-coupled receptors S1P1-5 (Hla and Dannenberg, 2012). However, some intracellular roles have been suggested for S1P, including the blocking of the histone deacetylases, HDAC1/2 (Hait et al., 2009) and the induction of apoptosis through interaction with BAK and BAX (Chipuk et al., 2012).
Recently, it was suggested that the RING domain of TRAF2 requires S1P as a co-factor for its E3 ligase activity (Alvarez et al., 2010). Alvarez and colleagues proposed that SPHK1 but not SPHK2 is activated by TNF and phosphorylates sphingosine to S1P which in turn binds to the RING domain of TRAF2 and serves as an essential co-factor that was missing in the experiments of Yin et al. Alvarez and colleagues, observed that in the absence of SPHK1, TNF-induced NF-κB activation was completely abolished.
Although we know a lot about TRAF2, there are still important gaps particularly with regard to cell type specificity and in vivo function of TRAF2. Moreover, despite the claims that SPHK1 and its product, S1P, are required for TRAF2 to function as a ubiquitin ligase, the responses of Traf2-/- and Sphk1-/- cells to TNF were not compared. Therefore, we undertook an analysis of TRAF2 and SPHK1 function in TNF signalling in a number of different tissues.
Surprisingly, we found that neither TRAF2 nor SPHK1 are required for TNF mediated canonical NF-κB and MAPK signalling in macrophages. However, MEFs, murine dermal fibroblasts (MDFs) and keratinocytes required TRAF2 but not SPHK1 for full strength TNF signalling. In these cell types, absence of TRAF2 caused a delay in TNF-induced activation of NF-κB and MAPK, and sensitivity to killing by TNF was increased. Absence of TRAF2 in keratinocytes in vivo resulted in psoriasis-like epidermal hyperplasia and skin inflammation. Unlike TNF-dependent genetic inflammatory skin conditions, such as IKK2 epidermal knock-out (Pasparakis et al., 2002) and the cpdm mutant (Gerlach et al., 2011), the onset of inflammation was only delayed, and not prevented by deletion of TNF. This early TNF-dependent inflammation is caused by excessive apoptotic but not necroptotic cell death and could be prevented by deletion of Casp8. We observed constitutive activation of NIK and non-canonical NF-κB in Traf2-/- keratinocytes which caused production of inflammatory cytokines and chemokines. We were able to reverse this inflammatory phenotype by simultaneously deleting both Tnf and Nfkb2 genes. Our results highlight the important role TRAF2 plays to protect keratinocytes from cell death and to down-regulate inflammatory responses and support the idea that intrinsic defects in keratinocytes can initiate psoriasis-like skin inflammation.
Article Section Process Block
Peer review process
This article was published via eLife's original peer review and publishing model, which was phased out in 2023. eLife's editors accepted this article for publication after peer review and was subject to mandatory revision requests before it was published.
History
- Version of Record published
- Preprint posted
- Preprint posted
- Preprint posted
- Sent for peer review
Article Section Without Title
TNF Receptor Associated Factor 2 (TRAF2) is an adaptor protein that transduces signals following ligation of certain cytokine receptors including those binding TNF. It was first identified together with TRAF1 as a component of TNF receptor-2 and then TNF receptor-1 (TNFR1) signalling complexes (Rothe et al., 1994; Shu et al., 1996). TRAF2, like most other TRAFs, contains a RING domain, several zinc fingers, a TRAF-N, and a conserved TRAF-C domain which is responsible for oligomerisation and receptor binding through its MATH region (Takeuchi et al., 1996; Uren and Vaux, 1996).
RING domains are nearly always associated with ubiquitin E3 ligase activity (Shi and Kehrl, 2003) and TRAF2 can promote ubiquitylation of RIPK1 in TNFR1 signalling complexes (TNFR1-SC) (Wertz et al., 2004). However TRAF2 recruits E3 ligases such as cIAPs to TNFR1-SC and these have also been shown to be able to ubiquitylate RIPK1 and regulate TNF signalling (Dynek et al., 2010; Mahoney et al., 2008; Varfolomeev et al., 2008; Vince et al., 2009). This makes it difficult to unambiguously determine the role of the E3 ligase activity of TRAF2.
Activation of JNK and NF-κB by TNF is reduced in cells from Traf2-/- mice while only JNK signalling was affected in lymphocytes from transgenic mice that express a dominant negative (DN) form of TRAF2 that lacks the RING domain (Lee et al., 1997; Yeh et al., 1997). Traf2-/-Traf5-/- mouse embryonic fibroblasts (MEFs) have a pronounced defect in activation of NF-κB by TNF, suggesting that absence of TRAF2 can be compensated by TRAF5 (Tada et al., 2001). Although activation of NF-κB was restored in Traf2-/-Traf5-/- cells by re-expression of wild type TRAF2, it was not restored when the cells were reconstituted with TRAF2 point mutants that could not bind cIAPs (Vince et al., 2009; Zhang et al., 2010). These data, together with a wealth of different lines of evidence showing that cIAPs are critical E3 ligases required for TNF-induced canonical NF-κB (Blackwell et al., 2013; Haas et al., 2009; Silke, 2011), support the idea that the main function of TRAF2 in TNF-induced NF-κB is to recruit cIAPs to the TNFR1-SC. However, it remains possible that the RING of TRAF2 plays another function, such as in activating JNK and protecting cells from TNF-induced cell death (Vince et al., 2009; Zhang et al., 2010). Furthermore it has been shown that TRAF2 can K48-ubiquitylate caspase-8 to set the threshold for TRAIL or Fas induced cell death (Gonzalvez et al., 2012). Moreover, TRAF2 inhibits non-canonical NF-κB signalling (Grech et al., 2004; Zarnegar et al., 2008) and this function requires the RING domain of TRAF2 to induce proteosomal degradation of NIK (Vince et al., 2009). However, structural and in vitro analyses indicate that, unlike TRAF6, the RING domain of TRAF2 is unable to bind E2 conjugating enzymes (Yin et al., 2009), and is therefore unlikely to have intrinsic E3 ligase activity.
Sphingosine-1-phosphate (S1P) is a pleiotropic sphingolipid mediator that regulates proliferation, differentiation, cell trafficking and vascular development (Pitson, 2011). S1P is generated by sphingosine kinase 1 and 2 (SPHK1 and SPHK2) (Kohama et al., 1998; Liu et al., 2000). Extracellular S1P mainly acts by binding to its five G protein-coupled receptors S1P1-5 (Hla and Dannenberg, 2012). However, some intracellular roles have been suggested for S1P, including the blocking of the histone deacetylases, HDAC1/2 (Hait et al., 2009) and the induction of apoptosis through interaction with BAK and BAX (Chipuk et al., 2012).
Recently, it was suggested that the RING domain of TRAF2 requires S1P as a co-factor for its E3 ligase activity (Alvarez et al., 2010). Alvarez and colleagues proposed that SPHK1 but not SPHK2 is activated by TNF and phosphorylates sphingosine to S1P which in turn binds to the RING domain of TRAF2 and serves as an essential co-factor that was missing in the experiments of Yin et al. Alvarez and colleagues, observed that in the absence of SPHK1, TNF-induced NF-κB activation was completely abolished.
Although we know a lot about TRAF2, there are still important gaps particularly with regard to cell type specificity and in vivo function of TRAF2. Moreover, despite the claims that SPHK1 and its product, S1P, are required for TRAF2 to function as a ubiquitin ligase, the responses of Traf2-/- and Sphk1-/- cells to TNF were not compared. Therefore, we undertook an analysis of TRAF2 and SPHK1 function in TNF signalling in a number of different tissues.
Surprisingly, we found that neither TRAF2 nor SPHK1 are required for TNF mediated canonical NF-κB and MAPK signalling in macrophages. However, MEFs, murine dermal fibroblasts (MDFs) and keratinocytes required TRAF2 but not SPHK1 for full strength TNF signalling. In these cell types, absence of TRAF2 caused a delay in TNF-induced activation of NF-κB and MAPK, and sensitivity to killing by TNF was increased. Absence of TRAF2 in keratinocytes in vivo resulted in psoriasis-like epidermal hyperplasia and skin inflammation. Unlike TNF-dependent genetic inflammatory skin conditions, such as IKK2 epidermal knock-out (Pasparakis et al., 2002) and the cpdm mutant (Gerlach et al., 2011), the onset of inflammation was only delayed, and not prevented by deletion of TNF. This early TNF-dependent inflammation is caused by excessive apoptotic but not necroptotic cell death and could be prevented by deletion of Casp8. We observed constitutive activation of NIK and non-canonical NF-κB in Traf2-/- keratinocytes which caused production of inflammatory cytokines and chemokines. We were able to reverse this inflammatory phenotype by simultaneously deleting both Tnf and Nfkb2 genes. Our results highlight the important role TRAF2 plays to protect keratinocytes from cell death and to down-regulate inflammatory responses and support the idea that intrinsic defects in keratinocytes can initiate psoriasis-like skin inflammation.
Assessment
- Landmark
- Fundamental
- Important
- Valuable
- Useful
- Exceptional
- Compelling
- Convincing
- Solid
- Incomplete
- Inadequate
During the peer-review process the editor and reviewers write an eLife Assessment that summarises the significance of the findings reported in the article (on a scale ranging from landmark to useful) and the strength of the evidence (on a scale ranging from exceptional to inadequate). Learn more about eLife Assessments
Call To Action

Captioned Asset
[Do not use this base pattern directly: see the following variants of this pattern for usage]
Captioned Asset Iframe Google Map
Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
Captioned Asset Iframe
Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
Captioned Asset Image
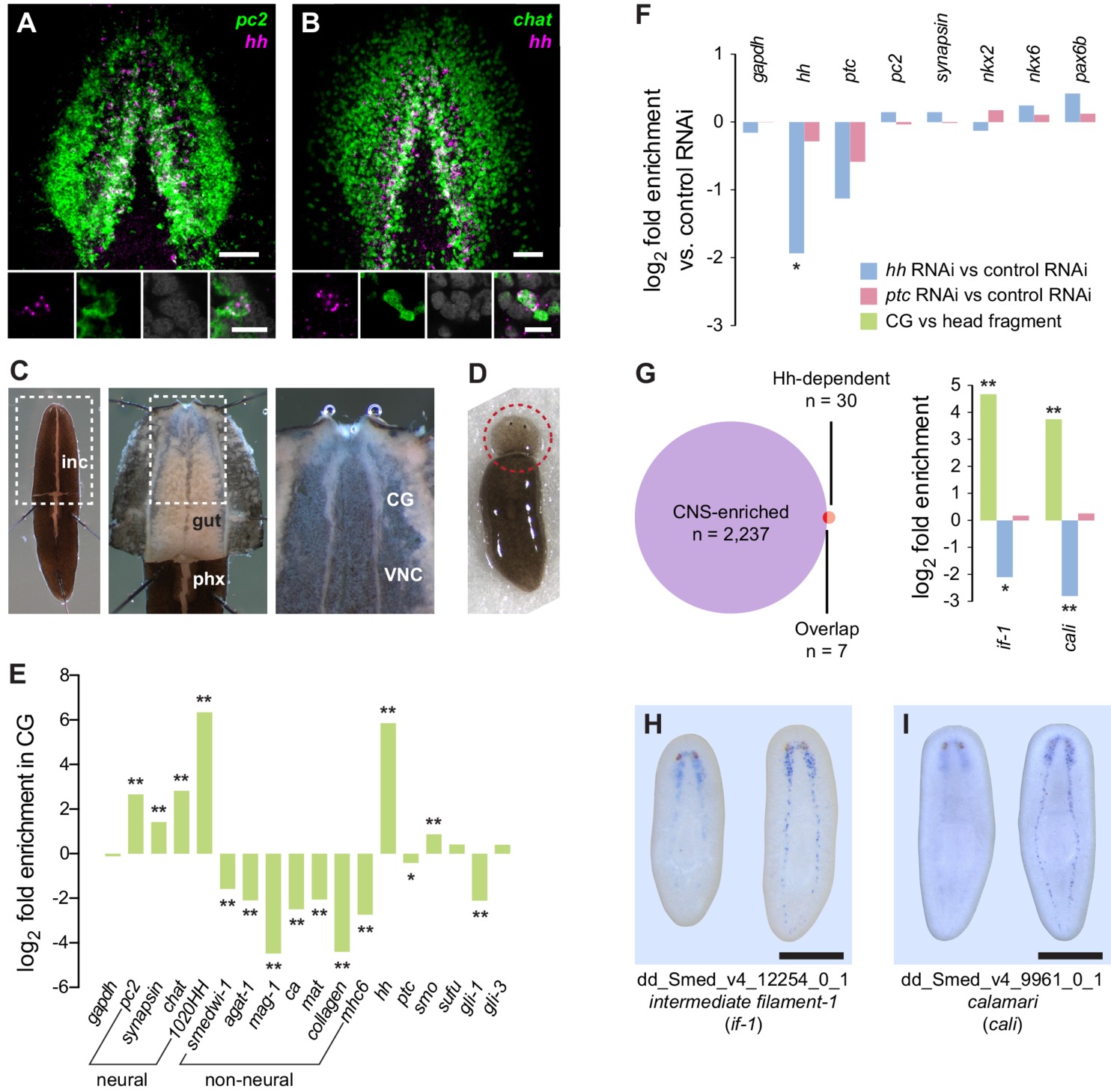
Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
Captioned Asset Inline Image

Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
Captioned Asset Picture

Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
Captioned Asset Table
Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
Captioned Asset Tables
Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
Captioned Asset Tweet
Tune in to the latest #eLifePodcast to hear authors discuss the stories behind their recent discoveries https://t.co/awwqu5L3lE
eLife - the journal (@eLife)
[Do not use this base pattern directly: see the following variants of this pattern for usage]
Captioned Asset Video
Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
Contextual Data
- Views 149
- Cited 5
- Annotations
Contextual Data With Citation
- Views 149
- Cited 5
- Annotations
Contextual Data With Download
- Views 149
- Downloads 20
- Cited 5
- Annotations
Highlight Item Blog Article

More eLife authors are linking submissions to their ORCID iDs
eLife sees positive results of requiring corresponding authors to register and link their profiles to their ORCID iDs
Highlight Item Event

eLife Continuum webinar
We're inviting all interested parties to participate in a webinar to learn about leveraging our new open-source publishing platform.
Highlight Item Press Pack

Scientists prove key aspect of evolutionary theory
While this event was first predicted almost twenty years ago, evidence for it has proved elusive.
Inline Profile

Lorem ipsum dolor sit amet, consectetur adipiscing elit. Mauris tempor euismod mi. Morbi facilisis eros in mi consequat cursus. Nulla malesuada pretium mauris, vitae gravida diam laoreet ac. Curabitur ornare libero sed porta molestie. Ut ut sem in libero tincidunt lobortis. Nullam eros nisi, malesuada vitae leo sit amet, bibendum congue justo. Morbi iaculis lectus erat, vel dapibus metus posuere sit amet. Phasellus eget scelerisque massa. Mauris hendrerit ex in nisl ultricies elementum. Donec tellus risus, rhoncus molestie rutrum et, cursus eget mi. Praesent et elit tellus. Curabitur id sodales turpis. Fusce pulvinar aliquam commodo. Cras vitae dolor vitae metus dignissim auctor.Cras nulla purus, rhoncus et mollis in, imperdiet sed metus. In non porttitor nibh. In sit amet ipsum eget nisl laoreet tincidunt. Donec iaculis nibh quis lacus viverra sagittis. Phasellus imperdiet congue imperdiet.
Interdum et malesuada fames ac ante ipsum primis in faucibus. Nunc nisi tortor, rhoncus.
Lorem ipsum dolor sit amet, consectetur adipiscing elit. Mauris tempor euismod mi. Morbi facilisis eros in mi consequat cursus. Nulla malesuada pretium mauris, vitae gravida diam laoreet ac. Curabitur ornare libero sed porta molestie. Ut ut sem in libero tincidunt lobortis. Nullam eros nisi, malesuada vitae leo sit amet, bibendum congue justo. Morbi iaculis lectus erat, vel dapibus metus posuere sit amet. Phasellus eget scelerisque massa. Mauris hendrerit ex in nisl ultricies elementum. Donec tellus risus, rhoncus molestie rutrum et, cursus eget mi. Praesent et elit tellus. Curabitur id sodales turpis. Fusce pulvinar aliquam commodo. Cras vitae dolor vitae metus dignissim auctor.Cras nulla purus, rhoncus et mollis in, imperdiet sed metus. In non porttitor nibh. In sit amet ipsum eget nisl laoreet tincidunt. Donec iaculis nibh quis lacus viverra sagittis. Phasellus imperdiet congue imperdiet.
Interdum et malesuada fames ac ante ipsum primis in faucibus. Nunc nisi tortor, rhoncus.
Media Chapter Listing Item
Mini Section
[heading]
[body]Personalised Cover Download
Download your personalised magazine cover
Thank you for publishing your research with us.
As a small keepsake for you and your co-authors we hope you’ll download this free poster highlighting your paper on an eLife magazine style cover.
Like your research it’s open access so you can print it, frame it, tweet it, use it as wallpaper in your lab...the possibilities are endless.
Profile Snippet
Pull Quote
Quote Incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur.
Excepteur sint est laborum.
Pull Quote With Cite
Quote Incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint est laborum.
May or may not be a link
Quote
Lorem ipsum dolor sit amet, consectetur adipiscing elit. Aenean interdum odio et sem fringilla, sit amet vulputate turpis sagittis. Cras non urna quam. Mauris vitae dui id ipsum varius semper. Curabitur sit amet viverra eros, ut maximus nisl. In hendrerit convallis turpis quis pulvinar. Donec porttitor faucibus ex, eget finibus velit vestibulum vitae. Donec nec nulla sollicitudin, volutpat enim interdum, sollicitudin erat.
Curabitur fermentum tellus ac rutrum efficitur. Phasellus libero magna, efficitur rutrum mauris a, mollis convallis lectus. Sed dapibus placerat eros ut vulputate. Vivamus quis sem accumsan, laoreet enim vel, dapibus ante. Nunc bibendum faucibus metus. Aliquam volutpat dapibus erat, quis ullamcorper nisl fermentum a. Praesent vulputate neque leo, vel rutrum dui auctor in. Duis eu magna et sem ullamcorper vehicula.
Quote With Cite
Lorem ipsum dolor sit amet, consectetur adipiscing elit. Aenean interdum odio et sem fringilla, sit amet vulputate turpis sagittis. Cras non urna quam. Mauris vitae dui id ipsum varius semper. Curabitur sit amet viverra eros, ut maximus nisl. In hendrerit convallis turpis quis pulvinar. Donec porttitor faucibus ex, eget finibus velit vestibulum vitae. Donec nec nulla sollicitudin, volutpat enim interdum, sollicitudin erat.
Curabitur fermentum tellus ac rutrum efficitur. Phasellus libero magna, efficitur rutrum mauris a, mollis convallis lectus. Sed dapibus placerat eros ut vulputate. Vivamus quis sem accumsan, laoreet enim vel, dapibus ante. Nunc bibendum faucibus metus. Aliquam volutpat dapibus erat, quis ullamcorper nisl fermentum a. Praesent vulputate neque leo, vel rutrum dui auctor in. Duis eu magna et sem ullamcorper vehicula.
May or may not be a link
Reference List
-
BookUnderstanding the rhythm of breathing: so near, yet so farIn: Julius D, Clapham DE, editors. Annual Review of Physiology 75:423–452.https://doi.org/10.1146/annurev-physiol-040510-130049
-
BookPhilosophie AnatomiqueParis: Méquignon-Marvis
-
Clinical TrialScripps Wired for Health Study (Registration: 000000)Scripps Translational Science Institute (sponsor) (2015)
-
ConferenceBoundary learning by optimization with topological constraintsBoundary learning by optimization with topological constraints, IEEE Conference on Computer Vision and Pattern Recognition (CVPR)https://doi.org/10.1038/365833a0
-
ConferenceBoundary learning by optimization with topological constraintsBoundary learning by optimization with topological constraints, IEEE Conference on Computer Vision and Pattern Recognition (CVPR)
-
ConferenceBoundary learning by optimization with topological constraintsBoundary learning by optimization with topological constraints, IEEE Conference on Computer Vision and Pattern Recognition (CVPR)
-
DataNKX2-5 mutations causative for congenital heart disease retain functionality and are directed to hundreds of targetsNCBI Gene Expression Omnibus. Series accession number GSE44902
-
PatentIRE-1alpha inhibitors (US20100941530)Mankind Corp, United States patent, United States
-
SoftwareThe Pymol Molecular Graphics SystemDeLano Scientific, Palo Alto, CA
Section Listing
Research categories
- Biochemistry
- Biophysics and structural biology
- Cancer biology
- Cell biology
- Computational and systems biology
- Developmental biology and stem cells
- Ecology
- Epidemiology and Global Health
- Genes and chromosomes
- Genomics and evolutionary biology
- Immunology
- Medicine
- Microbiology and infectious disease
- Neuroscience
- Plant biology
Section Listing Home Page
Categories
- Biochemistry
- Biophysics and structural biology
- Cancer biology
- Cell biology
- Computational and systems biology
- Developmental biology and stem cells
- Ecology
- Epidemiology and Global Health
- Genes and chromosomes
- Genomics and evolutionary biology
- Immunology
- Medicine
- Microbiology and infectious disease
- Neuroscience
- Plant biology
Section Listing Single
Magazine sections
Back to topSort Control
Statistic
- 320
- Downloads
Statistic Not Escaped Term
- 320
-
Citations for
version 1
Testimonial With Link

We liked the idea of having an open 'conversation' with the reviewers during the process, and having the chance to polish the manuscript by following the editors and reviewers’ recommendations without the threat of rejection.
Patrick Allard, Professor at University of California, Los Angeles
Why expert researchers support the eLife modelAsset Viewer Inline
Asset Viewer Inline Main Figure Nojs

Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
Asset Viewer Inline Main Figure With Supplement And Additional Assets Nojs

Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
How should this optional heading be styled?
-
Table 1 - source data 1.
Is the species name mentioned in the title, impact statement, abstract and digest of eLife papers
some more details here - https://doi.org/10.7554/eLife.10181.001
-
Table 1 - source data 1.
Is the species name mentioned in the title, impact statement, abstract and digest of eLife papers
some more details here - https://doi.org/10.7554/eLife.10181.002
Asset Viewer Inline Main Figure With Supplement And Additional Assets

Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
How should this optional heading be styled?
-
Table 1 - source data 1.
Is the species name mentioned in the title, impact statement, abstract and digest of eLife papers
some more details here - https://doi.org/10.7554/eLife.10181.001
-
Table 1 - source data 1.
Is the species name mentioned in the title, impact statement, abstract and digest of eLife papers
some more details here - https://doi.org/10.7554/eLife.10181.002
Asset Viewer Inline Supplementary Figure Nojs

Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
Asset Viewer Inline Supplementary Figure With Additional Assets Nojs

Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
How should this optional heading be styled?
-
Table 1 - source data 1.
Is the species name mentioned in the title, impact statement, abstract and digest of eLife papers
some more details here - https://doi.org/10.7554/eLife.10181.001
-
Table 1 - source data 1.
Is the species name mentioned in the title, impact statement, abstract and digest of eLife papers
some more details here - https://doi.org/10.7554/eLife.10181.002
How should this optional heading be styled?
-
Table 1 - source data 1.
Is the species name mentioned in the title, impact statement, abstract and digest of eLife papers
some more details here - https://doi.org/10.7554/eLife.10181.001
-
Table 1 - source data 1.
Is the species name mentioned in the title, impact statement, abstract and digest of eLife papers
some more details here - https://doi.org/10.7554/eLife.10181.002
Asset Viewer Inline Table Nojs
Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
Asset Viewer Inline Tables Nojs
Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
Asset Viewer Inline Video Nojs
Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
About Profiles
Leadership team
-
Randy Scheckman
Editor-in-Chief
Lorem ipsum dolor sit amet, consectetur adipiscing elit. Mauris tempor euismod mi. Morbi facilisis eros in mi consequat cursus. Nulla malesuada pretium mauris, vitae gravida diam laoreet ac. Curabitur ornare libero sed porta molestie. Ut ut sem in libero tincidunt lobortis. Nullam eros nisi, malesuada vitae leo sit amet, bibendum congue justo. Morbi iaculis lectus erat, vel dapibus metus posuere sit amet. Phasellus eget scelerisque massa. Mauris hendrerit ex in nisl ultricies elementum. Donec tellus risus, rhoncus molestie rutrum et, cursus eget mi. Praesent et elit tellus. Curabitur id sodales turpis. Fusce pulvinar aliquam commodo. Cras vitae dolor vitae metus dignissim auctor.Cras nulla purus, rhoncus et mollis in, imperdiet sed metus. In non porttitor nibh. In sit amet ipsum eget nisl laoreet tincidunt. Donec iaculis nibh quis lacus viverra sagittis. Phasellus imperdiet congue imperdiet.
Interdum et malesuada fames ac ante ipsum primis in faucibus. Nunc nisi tortor, rhoncus.
- Expertise
- Cell Biology
- Competing interests statement
- Donec tellus
-
Eve Marder
Deputy Editor
Lorem ipsum dolor sit amet, consectetur adipiscing elit. Mauris tempor euismod mi. Morbi facilisis eros in mi consequat cursus. Nulla malesuada pretium mauris, vitae gravida diam laoreet ac. Curabitur ornare libero sed porta molestie. Ut ut sem in libero tincidunt lobortis. Nullam eros nisi, malesuada vitae leo sit amet, bibendum congue justo. Morbi iaculis lectus erat, vel dapibus metus posuere sit amet. Phasellus eget scelerisque massa. Mauris hendrerit ex in nisl ultricies elementum. Donec tellus risus, rhoncus molestie rutrum et, cursus eget mi. Praesent et elit tellus. Curabitur id sodales turpis. Fusce pulvinar aliquam commodo. Cras vitae dolor vitae metus dignissim auctor.Cras nulla purus, rhoncus et mollis in, imperdiet sed metus. In non porttitor nibh. In sit amet ipsum eget nisl laoreet tincidunt. Donec iaculis nibh quis lacus viverra sagittis. Phasellus imperdiet congue imperdiet.
Interdum et malesuada fames ac ante ipsum primis in faucibus. Nunc nisi tortor, rhoncus.
- Expertise
- Cell Biology
- Competing interests statement
- Donec tellus
-
Profile (no image)
Deputy Editor
Lorem ipsum dolor sit amet, consectetur adipiscing elit. Mauris tempor euismod mi. Morbi facilisis eros in mi consequat cursus. Nulla malesuada pretium mauris, vitae gravida diam laoreet ac. Curabitur ornare libero sed porta molestie. Ut ut sem in libero tincidunt lobortis. Nullam eros nisi, malesuada vitae leo sit amet, bibendum congue justo. Morbi iaculis lectus erat, vel dapibus metus posuere sit amet. Phasellus eget scelerisque massa. Mauris hendrerit ex in nisl ultricies elementum. Donec tellus risus, rhoncus molestie rutrum et, cursus eget mi. Praesent et elit tellus. Curabitur id sodales turpis. Fusce pulvinar aliquam commodo. Cras vitae dolor vitae metus dignissim auctor.Cras nulla purus, rhoncus et mollis in, imperdiet sed metus. In non porttitor nibh. In sit amet ipsum eget nisl laoreet tincidunt. Donec iaculis nibh quis lacus viverra sagittis. Phasellus imperdiet congue imperdiet.
Interdum et malesuada fames ac ante ipsum primis in faucibus. Nunc nisi tortor, rhoncus.
- Expertise
- Cell Biology
- Competing interests statement
- Donec tellus
About Profiles Compact
Reviewing editors
-
Editor name
University of California, Los Angeles (United States)
- Research focus
- Pre-mRNA splicing
- post-transcriptional gene regulation
- Experimental organism
- Human
- Mouse
-
Editor name
University of California, Los Angeles (United States)
-
Editor name
University of California, Los Angeles (United States)
- Research focus
- Pre-mRNA splicing
- post-transcriptional gene regulation
- Experimental organism
- Human
- Mouse
-
Editor name
University of California, Los Angeles (United States)
- Research focus
- Pre-mRNA splicing
- post-transcriptional gene regulation
- Experimental organism
- Human
- Mouse
Additional Assets
How should this optional heading be styled?
-
Table 1 - source data 1.
Is the species name mentioned in the title, impact statement, abstract and digest of eLife papers (July–Sept 2014)?
some more details here - https://example.com/a-non-doi-link
-
Table 1 - source data 1.
Is the species name mentioned in the title, impact statement, abstract and digest of eLife papers (July–Sept 2014)?
some more details here - https://doi.org/10.7554/eLife.10181.001
Box
Widespread connectivity is not dependent on instrumentation and analyses.
This is an h1
This is an h2
This is an h3
This is an h4
This is an h5
This is an h6
This is a paragraph. It's a bit longer that the other things in this pattern so you can see how the wrapping works, which you can't really do unless you've got enough content. Right, that should do it. Onward!

Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
How should this optional heading be styled?
Decision Letter Header
eLife posts the editorial decision letter and author response on a selection of the published articles (subject to the approval of the authors). An edited version of the letter sent to the authors after peer review is shown, indicating the substantive concerns or comments; minor concerns are not usually shown. Reviewers have the opportunity to discuss the decision before the letter is sent (see review process). Similarly, the author response typically shows only responses to the major concerns raised by the reviewers.
Decision Letter Header With Profiles
-
Oliver HobertReviewing EditorColombia University, USA
-
Oliver HobertReviewing EditorColombia University, USA
eLife posts the editorial decision letter and author response on a selection of the published articles (subject to the approval of the authors). An edited version of the letter sent to the authors after peer review is shown, indicating the substantive concerns or comments; minor concerns are not usually shown.
Reviewers have the opportunity to discuss the decision before the letter is sent (see review process). Similarly, the author response typically shows only responses to the major concerns raised by the reviewers.
Filter Panel
Refine your results by:
Highlight Full
Highlights
-
-
Controlling traffic
Ramanath Hegde is a Postdoctoral Fellow at the Institute of Protein Biochemistry in Naples, Italy, where he investigates ways of preventing cells from destroying mutant proteins.
-
Controlling traffic
Ramanath Hegde is a Postdoctoral Fellow at the Institute of Protein Biochemistry in Naples, Italy, where he investigates ways of preventing cells from destroying mutant proteins.
Highlight One Item
Highlights
Highlight Six Items
Highlights
-
Infection, statistics and public health
Ramanath Hegde is a Postdoctoral Fellow at the Institute of Protein Biochemistry in Naples, Italy, where he investigates ways of preventing cells from destroying mutant proteins.
-
Infection, statistics and public health
Ramanath Hegde is a Postdoctoral Fellow at the Institute of Protein Biochemistry in Naples, Italy, where he investigates ways of preventing cells from destroying mutant proteins.
-
-
-
Controlling traffic
Ramanath Hegde is a Postdoctoral Fellow at the Institute of Protein Biochemistry in Naples, Italy, where he investigates ways of preventing cells from destroying mutant proteins.
-
Controlling traffic
Ramanath Hegde is a Postdoctoral Fellow at the Institute of Protein Biochemistry in Naples, Italy, where he investigates ways of preventing cells from destroying mutant proteins.
Listing Profile Snippets
Listing of profile snippets
-
Prabhat JhaUniversity of Toronto
-
Richard LosickHarvard University
- See more things!
Listing Read More
Further reading
-
- Cell Biology
- Epidemiology and Global Health
Optimal decision-making requires balancing fast but error-prone and more accurate but slower decisions through adjustments of decision thresholds. Here, we demonstrate two distinct correlates of such speed-accuracy adjustments by recording subthalamic nucleus (STN) activity and electroencephalography in 11 Parkinson’s disease patients during a perceptual decision-making task; STN low-frequency oscillatory (LFO) activity (2–8 Hz), coupled to activity at prefrontal electrode Fz, and STN beta activity (13–30 Hz) coupled to electrodes C3/C4 close to motor cortex. These two correlates differed not only in their cortical topography and spectral characteristics but also in the relative timing of recruitment and in their precise relationship with decision thresholds. Increases of STN LFO power preceding the response predicted increased thresholds only after accuracy instructions, while cue-induced reductions of STN beta power decreased thresholds irrespective of instructions. These findings indicate that distinct neural mechanisms determine whether a decision will be made in haste or with caution.
-
- Cell Biology
- Epidemiology and Global Health
Optimal decision-making requires balancing fast but error-prone and more accurate but slower decisions through adjustments of decision thresholds.
Listing Read More First Page
Further reading
-
- Cell Biology
- Epidemiology and Global Health
Optimal decision-making requires balancing fast but error-prone and more accurate but slower decisions through adjustments of decision thresholds. Here, we demonstrate two distinct correlates of such speed-accuracy adjustments by recording subthalamic nucleus (STN) activity and electroencephalography in 11 Parkinson’s disease patients during a perceptual decision-making task; STN low-frequency oscillatory (LFO) activity (2–8 Hz), coupled to activity at prefrontal electrode Fz, and STN beta activity (13–30 Hz) coupled to electrodes C3/C4 close to motor cortex. These two correlates differed not only in their cortical topography and spectral characteristics but also in the relative timing of recruitment and in their precise relationship with decision thresholds. Increases of STN LFO power preceding the response predicted increased thresholds only after accuracy instructions, while cue-induced reductions of STN beta power decreased thresholds irrespective of instructions. These findings indicate that distinct neural mechanisms determine whether a decision will be made in haste or with caution.
-
- Cell Biology
- Epidemiology and Global Health
Optimal decision-making requires balancing fast but error-prone and more accurate but slower decisions through adjustments of decision thresholds.
Read More Item
- Cell Biology
- Epidemiology and Global Health
Optimal decision-making requires balancing fast but error-prone and more accurate but slower decisions through adjustments of decision thresholds. Here, we demonstrate two distinct correlates of such speed-accuracy adjustments by recording subthalamic nucleus (STN) activity and electroencephalography in 11 Parkinson’s disease patients during a perceptual decision-making task; STN low-frequency oscillatory (LFO) activity (2–8 Hz), coupled to activity at prefrontal electrode Fz, and STN beta activity (13–30 Hz) coupled to electrodes C3/C4 close to motor cortex. These two correlates differed not only in their cortical topography and spectral characteristics but also in the relative timing of recruitment and in their precise relationship with decision thresholds. Increases of STN LFO power preceding the response predicted increased thresholds only after accuracy instructions, while cue-induced reductions of STN beta power decreased thresholds irrespective of instructions. These findings indicate that distinct neural mechanisms determine whether a decision will be made in haste or with caution.
Statistic Collection
-
- 54,741
- views
-
- 320
- downloads
-
- 89
- citations
Content Header Journal
[Do not use this base pattern directly: see the following variants of this pattern for usage]
- CommentOpen annotations (there are currently 0 annotations on this page).
Content Header Journal Article
Homo naledi, a new species of the genus Homo from the Dinaledi Chamber, South Africa
Content Header Journal Digest
Holding the focus
Content Header Journal Has Aside
Homo naledi, a new species of the genus Homo from the Dinaledi Chamber, South Africa
Content Header Journal Inside Elife Article
2022 eLife Community Ambassadors: Ready for action
Content Header Journal Magazine
Genetic Engineering: Increasing the uptake of carbon dioxide
Content Header Profile
Kananinoheaokuuhomeopuukaimanaalohilo
- University of West of Nowhere
- University of Middle of Nowhere
- University of East of Nowhere
Content Header Read More
- Cell Biology
- Epidemiology and Global Health
Content Header Simple
Browse our latest research
Content Header
Alerts and feeds
Content Header Background With Audio Player

Epidemiology and Global Health
Content Header Background With Image Credit And Audio Player

Epidemiology and Global Health
Content Header Background With Image Credit Overlay

Epidemiology and Global Health
Content Header Background With Image Credit

Epidemiology and Global Health
Content Header Background

Epidemiology and Global Health
Content Header Collection Title 0 Xx Short
Magazine
Content Header Collection Title 1 X Short

Scientist and Parent
Content Header Collection Title 2 Short

Reproducibility Project: Cancer Biology
Content Header Collection Title 3 Medium A

Reproducibility Project: Cancer Biology
Content Header Collection Title 3 Medium B

Mechanistic Microbiome Studies: A Special Issue
Content Header Collection Title 4 Long

eLife's Multi-format Plain-language Summaries of Research
Content Header Collection Title 5 X Long A

Ezrin enrichment on curved membranes requires a specific conformation or interaction with a curvature-sensitive partner
Content Header Collection Title 5 X Long B

Kasugamycin potentiates rifampicin and limits emergence of resistance in Mycobacterium tuberculosis by specifically decreasing mycobacterial mistranslation
Content Header Collection Title 6 Xx Long




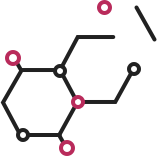








![Carbon Neutral Organisation carbon footprint standard [TM]](../../assets/img/patterns/molecules/carbon-neutral-organisation.png)










